Downloads
| Human Skin, Gene Catalog Reference[1] |
nucleotide sequences
fasta:pigGut_7.7M.GeneSet.fa.gz
amino acid sequences
fasta:pigGut_7.7M.GeneSet.pep.gz
|
| Integrated Human Skin Bacteria Genome Catalog[2] |
genomic DNA
https://db.cngb.org/search/organism/539655/
|
| genome catalog of the early-life human skin microbiome[3] |
raw metagenomic sequencing data
https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA971252
MAGs of the ELSG catalog
https://research.nhgri.nih.gov/projects/ELSG/downloads.shtml
|
| Human skin metagenome(Gene-Environment Interactions at the Skin Surface)[4] | |
| skin microbiome atopic dermatitis (AD)[5] |
sequencing data
https://www.ncbi.nlm.nih.gov/sra?term=SRP056821
|
| HMP project[6] | |
| Integrating cultivation and metagenomics for skin microbiome [7] |
Metagenome sequence data
https://www.ncbi.nlm.nih.gov/sra/?term=SRP002480
Metagenome assemblies, isolate and metagenome-assembled
genomes from the SMGC and genome annotations are available in:
https://ftp.ebi.ac.uk/pub/databases/metagenomics/genome_sets/skin_microbiome/
|
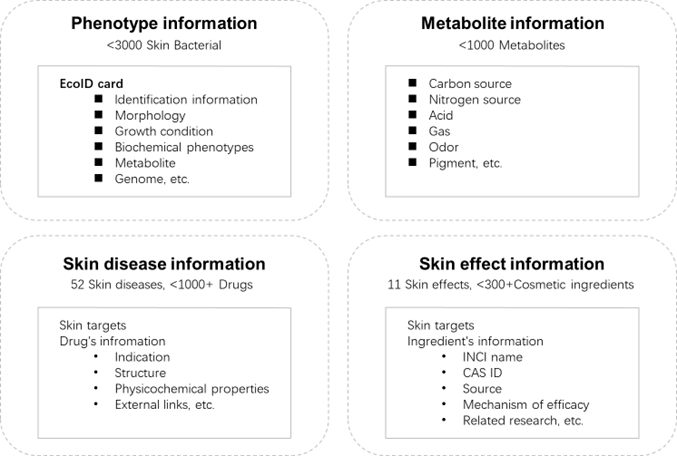
➢ Phenotypic information of organisms: Collected from original journal articles and classical textbooks (Bergey's Manual of Systematic Bacteriology and InternationalJournalofSystematicandEvolutionaryMicrobiology), combined with large language model (LLM) information extraction and manual review by domain experts to overcome data structuring challenges and ensure accuracy, then stored in an SQL database.
➢ Metabolite information: Gathered and organized microbial-related metabolite data by calling the PubMed database API.
➢ Disease and drug information: Compiled relevant data from the FAERS database (2014-2024).
➢ Skin efficacy and cosmetic ingredient information: Retrieved through PubMed searches using the keywords "skin effect" + "cosmetic ingredients" + "microorganisms", supplemented by manual curation.
✓ Prof. Xia Jingjing: xiajingjing@fudan.edu.cn
✓ Prof. Han Lianyi: hanlianyi@fudan.edu.cn
[1] Li Z, Xia J, Jiang L, et al. Characterization of the human skin resistome and identification of two microbiota cutotypes. Microbiome. 2021;9(1):47.
[2] Li Z, Ju Y, Xia J, et al. Integrated Human Skin Bacteria Genome Catalog Reveals Extensive Unexplored Habitat-Specific Microbiome Diversity and Function. Adv Sci (Weinh). 2023;10(28):e2300050.
[3] Shen Z, Robert L, Stolpman M, et al. A genome catalog of the early-life human skin microbiome. Genome Biol. 2023;24(1):252.
[4] Oh, J., Byrd, A., Deming, C. et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014).
[5] Chng, K., Tay, A., Li, C. et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol 1, 16106 (2016).
[6] The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
[7] Saheb Kashaf, S., Proctor, D.M., Deming, C. et al. Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nat Microbiol 7, 169–179 (2022).